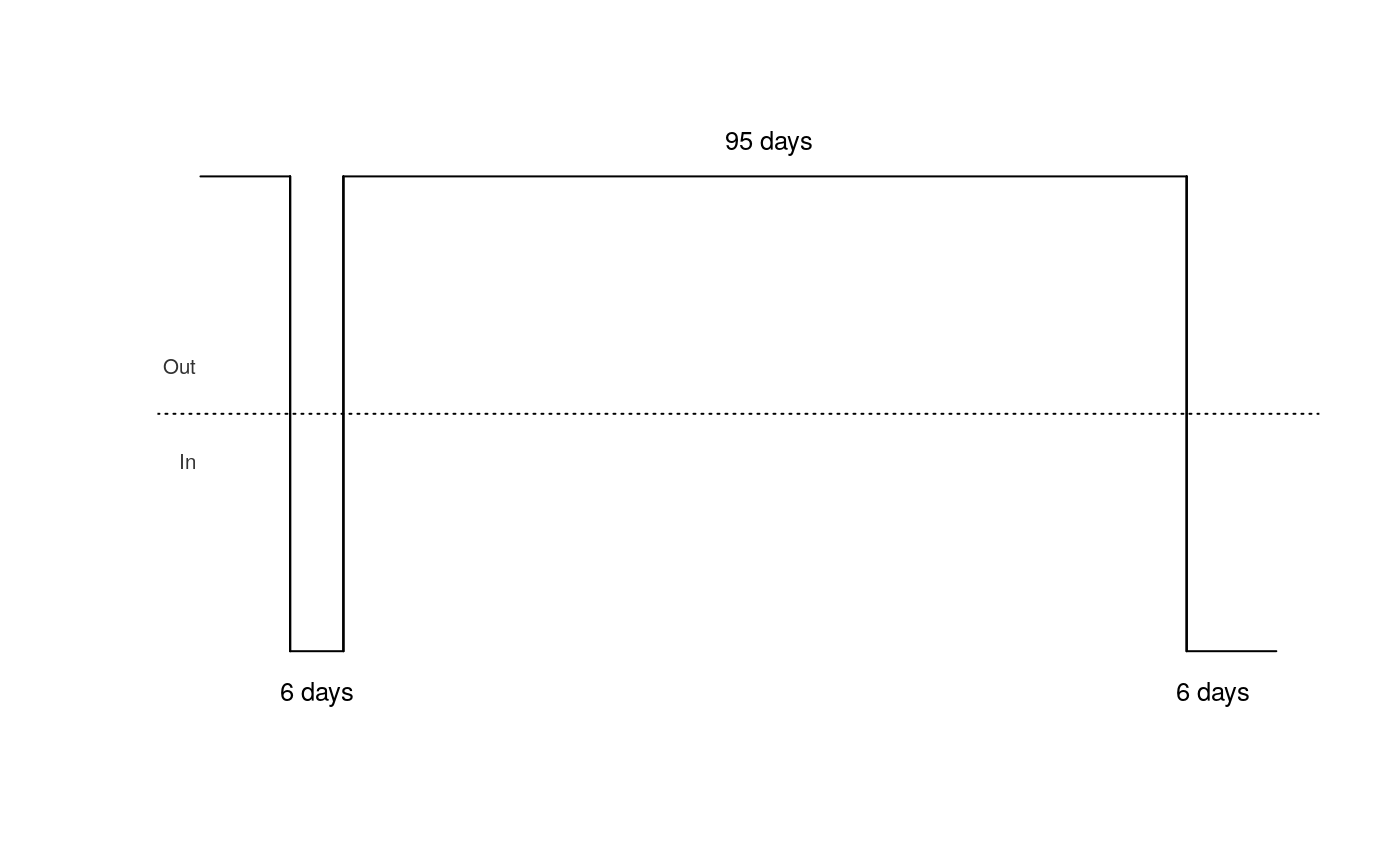
Given a sequence of border crossings for a person, draw a diagram describing that person's migration history.
Note that, unlike elsewhere in package migrbc, the
date_crossing and is_arrival arguments for
plot_mig_hist refer to a single individual.
If values for date_first and date_last are not supplied,
then defaults are calculated, based on the length of the travel history.
plot_mig_hist(date_crossing, is_arrival, days_to_next_crossing, res_status_before_str = NULL, res_status_after_str = NULL, date_first = NULL, date_last = NULL, show_dates = TRUE, show_days = TRUE, cex = 1, lwd = 1)
Arguments
| date_crossing | A vector of dates. |
|---|---|
| is_arrival | A logical vector, the same length as |
| days_to_next_crossing | A number vector, the same length as
|
| res_status_before_str | Character or numeric vector,
the same length as |
| res_status_after_str | Character or numeric vector,
the same length as |
| date_first | The start date for the travel history. Optional. |
| date_last | The end date for the travel history. Optional. |
| show_dates | Logical. Whether to display the dates of each border crossing. |
| show_days | Logical. Whether to display the length, in days, of each spell in or out of the country. |
| cex | 'Character expansion factor'. A number. Larger values lead to larger text. Defaults to 1. |
| lwd | Line width. A number. Larger values lead to thicker lines. Defaults to 1. |
Value
Returns NULL, but as a side effect draws a graph
(using R's traditional graphics system).
Examples
#> logger 'migrbc' has log_level=1 and writes to the consoleplot_test <- function(mig_hist) { plot_mig_hist(date_crossing = as.character(mig_hist$date_crossing), is_arrival = mig_hist$is_arrival, days_to_next_crossing = mig_hist$days_to_next_crossing, show_date = FALSE, cex = 0.8) } number_of_people = 1 person_data <- migrbc::setup_random_test_data(number_of_people, initial_date = '2001-01-01', numJourneys = 3, min = 0, max = 100) cross_spaces <- migrbc::pre_process(person_data, n_groups = 1) ## run in non-parallel post_data <- migrbc::run_rbc(cross_spaces, window_size = 487, threshold_year = 365, parallel=FALSE) old_par <- par(mfrow = c(1, 1)) plot_test(post_data$journeys)par(old_par)